Cheng Zeng
Postdoctoral associate at University of Florida.

Phone: +1 401-396-6668
Email: c.zeng@ufl.edu
Hi! I’m currently a postdoctoral researcher at the University of Florida. Before joining UF, I was a postdoctoral fellow at the Institute of Experiential AI and Roux Institute of Northeastern University. I received my Ph.D. in Chemical Engineering and an M.S. in Data Science in 2022 from Brown University.
My research lies at the intersection of AI, Materials Science and Chemistry. I develop and deploy generative models, machine learning interatomic potentials, and phenomenological models to accelerate materials design for a sustainable future.
I’m currently on the job market—welcome to connect via LinkedIn!
Recent news [archive]
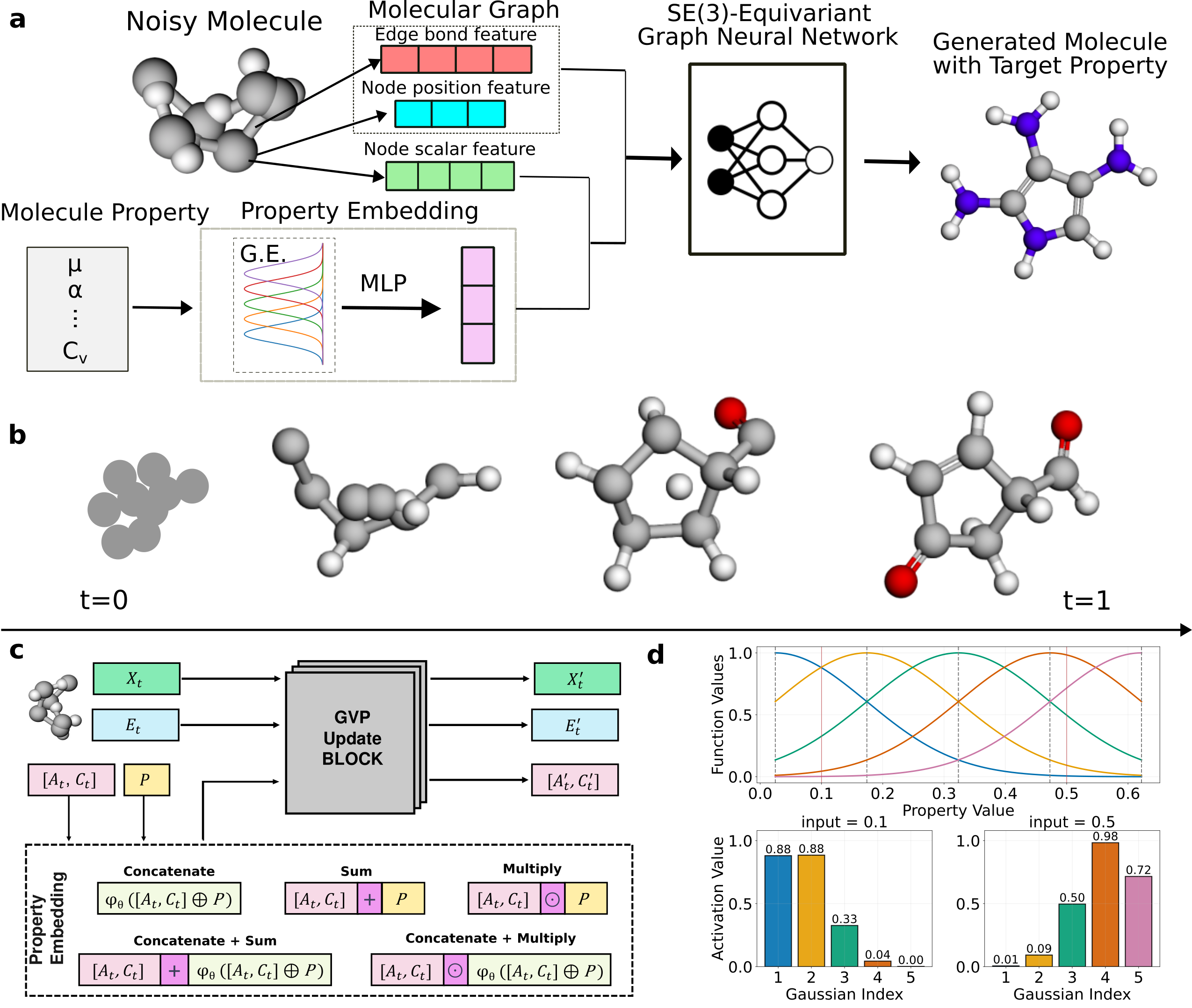
| 2026/01 | Our PropMolFlow work is finally online at Nature Computational Science. News release is here. |
|---|---|
| 2025/11 | Our team at the University of Florida, including Cheng Zeng, Jirui Jin, and Mingjie Liu, has been honored with the HiPerGator Pioneering Team Award for our work on developing advanced graph neural network techniques to explore the vast chemical space. This recognition, awarded by the UF Artificial Intelligence and Informatics Research Institute, highlights the impact of combining high-performance computing with cutting-edge AI for chemistry and materials discovery. The official news release can be found here. |
| 2025/07 | Our paper MolGuidance, a comprehensive extension to PropMolFlow, has been accepted in ICML 2025 Generative AI for Biology (GenBio) Workshop. In this work, we integrate three advanced property‐guidance strategies—classifier‐free guidance, autoguidance, and model guidance—into an SE(3)‐equivariant flow‐matching framework for conditional molecule generation. The dataset is available at zenodo. |